Compute and storage resources
Our group has access to considerable computation and storage resources. This document describes these and how to access them. Each resource has its own use scenario, and some may be more adapted to specific problems than others (for example, if you want to set up a Twitter crawler, use a Nectar Virtual Machine for better uptime and to avoid security issues on the compute servers). We have four compute resources and each has attached a storage resource.
Compute servers and frameworks
iHPC servers
Link to resource: ihpc.research.uts.edu.au (requires UTS VPN)
Quick description: iHPC stands for Interactive High-Performance Computing. In a nutshell, these are very large machines (around 60 cores, and terabytes of memory). The machines are hosted in UTS and require both a UTS affiliation and special access. The servers are grouped into clusters, some of which require special access.
Our group has a private cluster called hephaestos which currently contains three machines (see print screen below). Our private cluster is accessible to BDS members only.
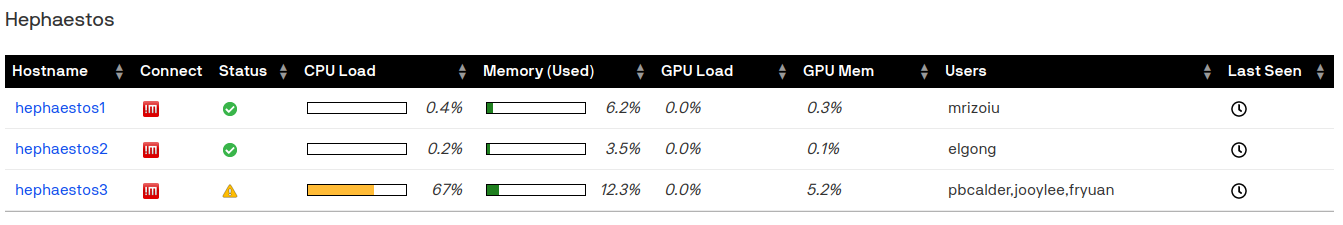
Getting access: Request an iHPC account (see ihpc.research.uts.edu.au for instructions). Mention the Behavioral Data Science group and Andrei in your request to be automatically added to the hephaestos access lists. Make sure you read the documentation!
Use case: the iHPC is designed for prototyping that requires large machines (Jupyter Notebook or RStudio), or for intermediary-size compute (for larger size see UTS HPC and NCI). You can use iHPC using its Graphical User Interface (GUI) via NoMachine. You can also connect via console using:
ssh <user>@hephaestos1.ihpc.uts.edu.au
For use with Jupyter Notebook or RStudio Server, you can redirect ports via SSH tunneling. Assuming the Jupyter Notebook/RStudio Server listens on port 8282, you can redirect that port by connecting to hephaestos1 using:
ssh -L 8282:localhost:8282 <user>@hephaestos1.ihpc.uts.edu.au
after which you can connect the browser to your local machine to https://localhost:8282 and access the kernel on the distant machines. See here for more details and examples.
More details: For detailed instructions on setting up remote Jupyter notebooks on iHPC, check out this guide.
NeCTAR Virtual Machines
Link to resource: https://dashboard.rc.nectar.org.au/
Quick description: NeCTAR is an infrastructure for virtual machines (i.e. machines that can be spawned for a particular purpose), similar to Amazon Web Services. This is quite useful for crawling data with no downtime, hosting visualizers, and even computing. More details here: https://ardc.edu.au/services/nectar-research-cloud/
Our group has an allocation of 25 instances, 120 vCPUs, and 480 GB of RAM.
Getting access: First activate your UTS/ANU/Data61 account here: https://dashboard.rc.nectar.org.au/. Then flip Andrei the email address associated with the account, and I’ll add you to the project. Note: once you have access, you need to switch projects using the project selector in the top-left corner to access the behavioral-ds allocation.
Use case: You can easily spawn virtual machines, install stuff on them, play and destroy them. Each virtual machine acts as a computer, therefore you can install web interfaces, use it for compute (the larger flavors), and connect via SSH. The port forwarding trick described for iHPC also works for NeCTAR VMs.
UTS High-Performance Computing Cluster (HPCC)
Link to resource: https://hpc.research.uts.edu.au/
Usage status: https://hpc.research.uts.edu.au/status/
Quick description: HPCC is a typical high-performance computing cluster (a.k.a. super-computers), in which jobs run non-interactively. See here a schema of HPC clusters. UTS HPCC uses PBS as the scheduler, and it is designed to be fully compatible with NCI. Its mission is to be a training or development site for larger HPC projects destined for NCI.
Getting access: You need to request access to the HPCC. Email eResearch-IT@uts.edu.au to introduce yourself and your requirements. Make sure you mention Andrei and that you are part of the Behavioral DS group. Once you have access read the HPC Getting Started pages.
Use case: As HPC systems stand, UTS HPCC is a small system, but very useful for mid- to largish-scale computations. You can access 600 cores and up to 6TB of memory. Usage-wise, UTS HPCC stands in between iHPC and the NCI. That is, once your compute grows too large for iHPC, you use HPCC to scale it up and prepare your scripts for NCI.
National Computation Infrastructure (NCI) supercomputer – Gadi
Link to resource: https://nci.org.au/
Usage status: https://nci.org.au/our-systems/status
Quick description: NCI’s Gadi is Australia’s supercomputer. Gadi has 3,200 compute nodes, 155,000 CPU cores, 567 Terabytes of memory, and 640 GPUs (as of 07/09/2021). It has a peak performance of 9 petaflops (by comparison, the largest supercomputer, Fugaku in Japan, has a peak performance of 442 petaflops).
Gadi is a cross-institutional shared resource, and its usage requires credit (in NCI terms service units (SU)). Our group has a varying allocation of SUs to be used for computing, which can be dynamically increased. Speak to Andrei if you believe you may require more than we have or even if you may use up all the exiting allocation (not leaving enough for the others).
Getting access: You will need to request an NCI account using your UTS/ANU/Data61 email address. Use our project code gh47: Tracking disinformation campaigns across social media (Marian-Andrei Rizoiu) during the joining process. If you already have an account, simply request to join the project. You will subsequently use this project code for all computing. Make sure you have a read of the Gadi User guide.
Use case: Use Gadi for the largest scale computes, which require years of sequential compute time. Ideally, you designed already your scripts using the UTS HPCC. Note that Gadi is less responsive than HPCC (and less forgiving with not respecting the limits and instructions).
There are several queues available for Gadi, depending on your usage requirements. These include normal (day-to-day compute), express (small batches that can be executed fast), hugemem and megamem (large memory requirement), and gpuvolta (for deep learning).
Storage
iHPC data folder (1TB)
iHPC home folders are limited to 32 GB. However, each iHPC user has a data folder of 1TB accessible at /data/<username>. Put here all large files, including your anaconda and datasets. This data folder is on Network File System (NFS) and accessible from any iHPC machine.
iHPC project mount (20TB)
For shared files and datasets, our group has a project allocation of 20TB, accessible at /projects/BehavioralDS/. All files in this folder are accessible by all group members.
Nectar storage (12TB)
NeCTAR VMs have very little storage space (~30GB of disk). You can also create external volumes that can be attached to the VMs (each 50GB). For larger storage, our group has a 12TB NFS volume that needs to be attached using the following procedure.
Send the IP address or hostname of the machine that you want to mount the Space storage to Intersect Operations (help@intersect.org.au). Copy Andrei to the email!
Install NFS and automount related packages (for Ubuntu):
apt-get install nfs-common autofs rpcbind
systemctl enable rpcbind
systemctl start rpcbind
systemctl enable autofs
systemctl start autofs
- Create Directories
mkdir -p /data/mounts
- Create /etc/auto.data
echo "Q3530 -fstype=nfs,nfsvers=3 10.255.122.28:/gpfs/general00/pool9000/Q3530/Q3530" >> /etc/auto.data
- Modify /etc/auto.master
Run the following command to insert mount point to /etc/auto.master file
echo "/data/mounts /etc/auto.data" >> /etc/auto.master
- Restart autofs and create symlink
Restart autofs service to apply the above changes
systemctl restart autofs
ln -s /data/mounts/Q3530 /data/Q3530
NCI storage (4TB)
Our group has a storage allocation of 4TB on NCI. It can be accessed at /g/data/gh47/. If you want to use this allocation in your PBS jobs, you need to explicitly ask for it in the submission script. For example, to indicate that credit should be used from gh47 and to access both the scratch and the /g/data allocation, one would add the following lines in the submission script:
#PBS -P gh47
#PBS -l storage=scratch/gh47+g/data/gh47